Evrards Collapse#
Imports#
# ==== GPU selection ====
from autocvd import autocvd
autocvd(num_gpus = 1)
# =======================
# numerics
import jax
import jax.numpy as jnp
# timing
from timeit import default_timer as timer
# plotting
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
# astronomix classes
from astronomix import SimulationConfig
from astronomix import SimulationParams
from astronomix.option_classes.simulation_config import BoundarySettings, BoundarySettings1D
# astronomix functions
from astronomix import get_helper_data
from astronomix import time_integration
from astronomix import construct_primitive_state
from astronomix.option_classes.simulation_config import finalize_config
from astronomix import get_registered_variables
# astronomix constants
from astronomix.option_classes.simulation_config import (
BACKWARDS, FORWARDS, HLL, HLLC, MINMOD, OSHER,
PERIODIC_BOUNDARY, REFLECTIVE_BOUNDARY,
BoundarySettings, BoundarySettings1D
)
Initiatization#
from astronomix.option_classes.simulation_config import DONOR_ACCOUNTING, DOUBLE_MINMOD, HLLC_LM, LAX_FRIEDRICHS, MUSCL, RIEMANN_SPLIT_UNSTABLE, SIMPLE_SOURCE_TERM, SPLIT, VAN_ALBADA, VAN_ALBADA_PP
print("👷 Setting up simulation...")
# simulation settings
gamma = 5/3
# spatial domain
box_size = 4.0
num_cells = 128
fixed_timestep = False
dt_max = 0.001
# setup simulation config
config = SimulationConfig(
runtime_debugging = False,
progress_bar = True,
self_gravity = True,
dimensionality = 3,
box_size = box_size,
num_cells = num_cells,
split = SPLIT,
time_integrator = MUSCL,
fixed_timestep = fixed_timestep,
riemann_solver = HLLC,
limiter = MINMOD,
self_gravity_version = RIEMANN_SPLIT_UNSTABLE,
differentiation_mode = FORWARDS,
boundary_settings = BoundarySettings(
BoundarySettings1D(
left_boundary = REFLECTIVE_BOUNDARY,
right_boundary = REFLECTIVE_BOUNDARY
),
BoundarySettings1D(
left_boundary = REFLECTIVE_BOUNDARY,
right_boundary = REFLECTIVE_BOUNDARY
),
BoundarySettings1D(
left_boundary = REFLECTIVE_BOUNDARY,
right_boundary = REFLECTIVE_BOUNDARY
)
)
)
helper_data = get_helper_data(config)
params = SimulationParams(
t_end = 0.8,
C_cfl = 0.4,
dt_max = dt_max,
)
registered_variables = get_registered_variables(config)
👷 Setting up simulation...
Setting the initial state#
from astronomix.fluid_equations.fluid import construct_primitive_state3D
R = 1.0
M = 1.0
dx = config.box_size / (config.num_cells - 1)
# initialize density field
num_injection_cells = jnp.sum(helper_data.r <= R)
rho = jnp.where(helper_data.r <= R, M / (2 * jnp.pi * R ** 2 * helper_data.r), 1e-4)
total_injected_mass = jnp.sum(jnp.where(helper_data.r <= R, rho, 0)) * dx ** 3
print(f"Injected mass: {total_injected_mass}")
# better ball edges
# overlap_weights = (R + dx / 2 - helper_data.r) / dx
# rho = jnp.where((helper_data.r > R - dx / 2) & (helper_data.r < R + dx / 2), rho * overlap_weights, rho)
# Initialize velocity fields to zero
v_x = jnp.zeros_like(rho)
v_y = jnp.zeros_like(rho)
v_z = jnp.zeros_like(rho)
# initial thermal energy per unit mass = 0.05
e = 0.05
p = (gamma - 1) * rho * e
# Construct the initial primitive state for the 3D simulation.
initial_state = construct_primitive_state(
config = config,
registered_variables = registered_variables,
density = rho,
velocity_x = v_x,
velocity_y = v_y,
velocity_z = v_z,
gas_pressure = p
)
# sharding
# actually leads to a slow down, to fix
# from jax.sharding import PartitionSpec as P, NamedSharding
# sharding_mesh = jax.make_mesh((1, 2, 2, 1), ('variables', 'x', 'y', 'z'))
# initial_state = jax.device_put(initial_state, NamedSharding(sharding_mesh, P('variables', 'x', 'y', 'z')))
Injected mass: 1.0016810894012451
config = finalize_config(config, initial_state.shape)
Consider using RIEMANN_SPLIT as the self_gravity_version.
Simulation#
final_state = time_integration(initial_state, config, params, registered_variables)
|████████████████████████████████████████████████████████████████████| 100.0%
Visualization#
Cut#
from matplotlib.colors import LogNorm
a = num_cells // 2 - 30
b = num_cells // 2 + 30
c = num_cells // 2 + 20
d = num_cells // 2 + 50
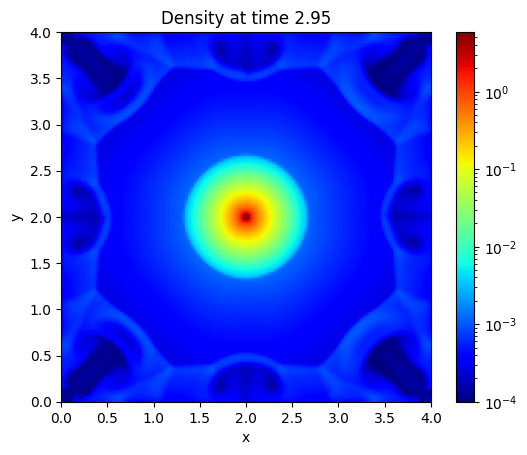
plt.imshow(jnp.abs(final_state[registered_variables.pressure_index, :, :, num_cells // 2].T), cmap = "jet", origin = "lower", extent=[0, box_size, 0, box_size], norm = LogNorm())
plt.colorbar()
plt.xlabel("x")
plt.ylabel("y")
Text(0, 0.5, 'y')
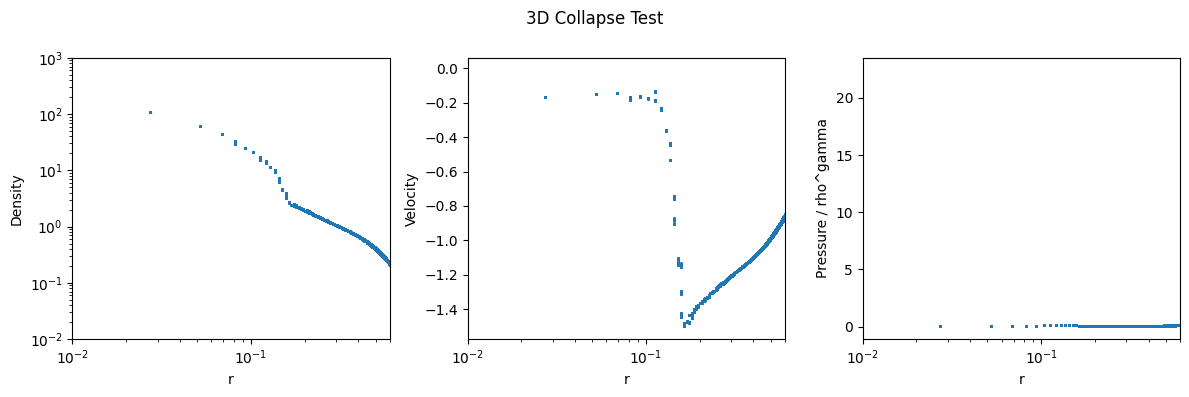
# plot the radial density profile rho over r in a log-log plot
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(12, 4))
ax1.scatter(helper_data.r.flatten(), final_state[registered_variables.density_index].flatten(), label="Final Density", s = 1)
# x and y log scale
ax1.set_xscale("log")
ax1.set_yscale("log")
ax1.set_xlim(1e-2, 6e-1)
ax1.set_ylim(1e-2, 1e3)
ax1.set_xlabel("r")
ax1.set_ylabel("Density")
# velocity profile
v_r = -jnp.sqrt(final_state[registered_variables.velocity_index.x] ** 2 + final_state[registered_variables.velocity_index.y] ** 2 + final_state[registered_variables.velocity_index.z] ** 2)
ax2.scatter(helper_data.r.flatten(), v_r.flatten(), label="Radial Velocity", s = 1)
# log x scale
ax2.set_xscale("log")
ax2.set_xlim(1e-2, 6e-1)
ax2.set_xlabel("r")
ax2.set_ylabel("Velocity")
# plot P / rho^gamma
ax3.scatter(helper_data.r.flatten(), final_state[registered_variables.pressure_index].flatten() / final_state[registered_variables.density_index].flatten() ** gamma, label="P / rho^gamma", s = 1)
# ax3.set_xlim(box_size / num_cells, 6e-1)
ax3.set_xlabel("r")
ax3.set_ylabel("Pressure / rho^gamma")
ax3.set_xscale("log")
ax3.set_xlim(1e-2, 6e-1)
fig.suptitle("3D Collapse Test")
plt.tight_layout()
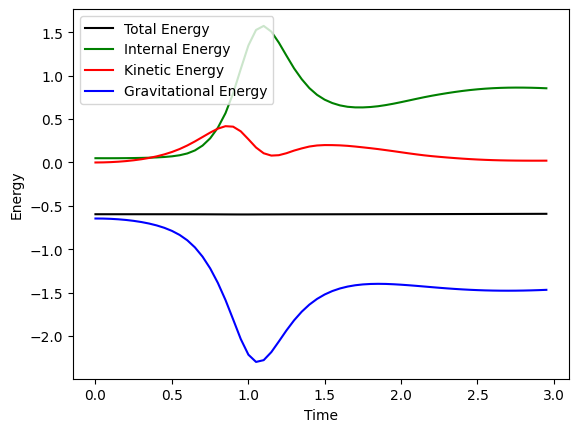
Conservational properties#
config = config._replace(return_snapshots = True, num_snapshots = 60)
params = params._replace(t_end = 3.0)
snapshots = time_integration(initial_state, config, params, registered_variables)
|████████████████████████████████████████████████████████████████████| 100.0%
total_energy = snapshots.total_energy
internal_energy = snapshots.internal_energy
kinetic_energy = snapshots.kinetic_energy
gravitational_energy = snapshots.gravitational_energy
total_mass = snapshots.total_mass
time = snapshots.time_points
t_end = 3.0
plt.plot(time, total_energy, label="Total Energy", color = "black")
plt.plot(time, internal_energy, label="Internal Energy", color = "green")
plt.plot(time, kinetic_energy, label="Kinetic Energy", color = "red")
plt.plot(time, gravitational_energy, label="Gravitational Energy", color = "blue")
plt.xlabel("Time")
plt.ylabel("Energy")
plt.legend()
plt.savefig("collapse_conservation.svg")
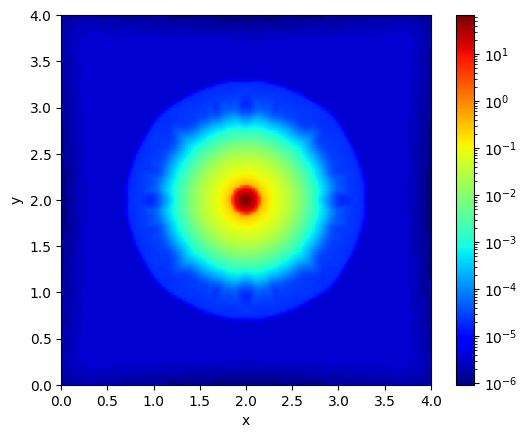
import matplotlib.animation as animation
fig, ax = plt.subplots()
def animate(i):
ax.clear()
state = snapshots.states[i]
im = ax.imshow(state[0, :, :, num_cells // 2].T, cmap="jet", origin="lower", extent=[0, box_size, 0, box_size], norm=LogNorm())
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.set_title(f"Density at time {time[i]:.2f}")
return [im]
ani = animation.FuncAnimation(fig, animate, frames=len(snapshots.states), interval=100, blit=True)
plt.colorbar(ax.imshow(snapshots.states[0][0, :, :, num_cells // 2].T, cmap="jet", origin="lower", extent=[0, box_size, 0, box_size], norm=LogNorm()), ax=ax)
# save to gif
ani.save("3d_collapse.gif")