Conservational Properties#
Imports#
# ==== GPU selection ====
from autocvd import autocvd
autocvd(num_gpus = 1)
# =======================
# numerics
import jax
import jax.numpy as jnp
from jax import grad
# 64-bit precision
jax.config.update("jax_enable_x64", True)
# plotting
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
# astronomix
from astronomix import SimulationConfig
from astronomix import get_helper_data
from astronomix import SimulationParams
from astronomix import time_integration
from astronomix import construct_primitive_state
from astronomix import get_registered_variables
from astronomix._physics_modules._stellar_wind.stellar_wind_options import WindConfig
from astronomix.option_classes.simulation_config import SPHERICAL
from astronomix.option_classes.simulation_config import finalize_config
Initiating the stellar wind simulation#
print("👷 Setting up simulation...")
# simulation settings
gamma = 5/3
# spatial domain
geometry = SPHERICAL
box_size = 1.0
num_cells = 101
fixed_timestep = False
return_snapshots = True
num_snapshots = 40
# activate stellar wind
stellar_wind = False
# setup simulation config
config = SimulationConfig(
geometry = geometry,
box_size = box_size,
num_cells = num_cells,
wind_config = WindConfig(
stellar_wind = stellar_wind,
),
fixed_timestep = fixed_timestep,
return_snapshots = return_snapshots,
num_snapshots = num_snapshots
)
helper_data = get_helper_data(config)
registered_variables = get_registered_variables(config)
👷 Setting up simulation...
Setting the simulation parameters and initial state#
# time domain
dt_max = 0.1
C_CFL = 0.8
t_end = 0.2
# SOD shock tube
shock_pos = 0.5
r = helper_data.geometric_centers
rho = jnp.where(r < shock_pos, 1.0, 0.125)
u = jnp.zeros_like(r)
p = jnp.where(r < shock_pos, 1.0, 0.1)
# get initial state
initial_state = construct_primitive_state(
config = config,
registered_variables = registered_variables,
density = rho,
velocity_x = u,
gas_pressure = p
)
params = SimulationParams(C_cfl = C_CFL, dt_max = dt_max, gamma = gamma, t_end = t_end)
Conservation test of the simulation#
def init_shock_problem(num_cells, config, params, first_order_fallback = False):
config = config._replace(num_cells = num_cells, grid_spacing = config.box_size / (num_cells - 1), first_order_fallback = first_order_fallback)
params = params._replace(dt_max = dt_max)
helper_data = get_helper_data(config)
r = helper_data.geometric_centers
rho = jnp.where(r < shock_pos, 1.0, 0.125)
u = jnp.zeros_like(r)
p = jnp.where(r < shock_pos, 1.0, 0.1)
# get initial state
initial_state = construct_primitive_state(
config = config,
registered_variables = registered_variables,
density = rho,
velocity_x = u,
gas_pressure = p
)
config = finalize_config(config, initial_state.shape)
return initial_state, config, params, helper_data
initial_state_shock100, config_shock100, params_shock100, helper_data_shock100 = init_shock_problem(101, config, params)
checkpoints_shock100 = time_integration(initial_state_shock100, config_shock100, params_shock100, registered_variables)
initial_state_shock100_first_order, config_shock100_first_order, params_shock100_first_order, helper_data_shock100_first_order = init_shock_problem(101, config, params, True)
checkpoints_shock100_first_order = time_integration(initial_state_shock100_first_order, config_shock100_first_order, params_shock100_first_order, registered_variables)
initial_state_shock, config_shock, params_shock, helper_data_shock = init_shock_problem(2001, config, params, True)
checkpoints_shock2000 = time_integration(initial_state_shock, config_shock, params_shock, registered_variables)
For spherical geometry, only HLL is currently supported. Also, only the unsplit mode has been tested.
Setting unsplit mode for spherical geometry
Setting MUSCL time integrator for spherical geometry
Automatically setting reflective left and open right boundary for spherical geometry.
For spherical geometry, only HLL is currently supported. Also, only the unsplit mode has been tested.
Setting unsplit mode for spherical geometry
Setting MUSCL time integrator for spherical geometry
Automatically setting reflective left and open right boundary for spherical geometry.
For spherical geometry, only HLL is currently supported. Also, only the unsplit mode has been tested.
Setting unsplit mode for spherical geometry
Setting MUSCL time integrator for spherical geometry
Automatically setting reflective left and open right boundary for spherical geometry.
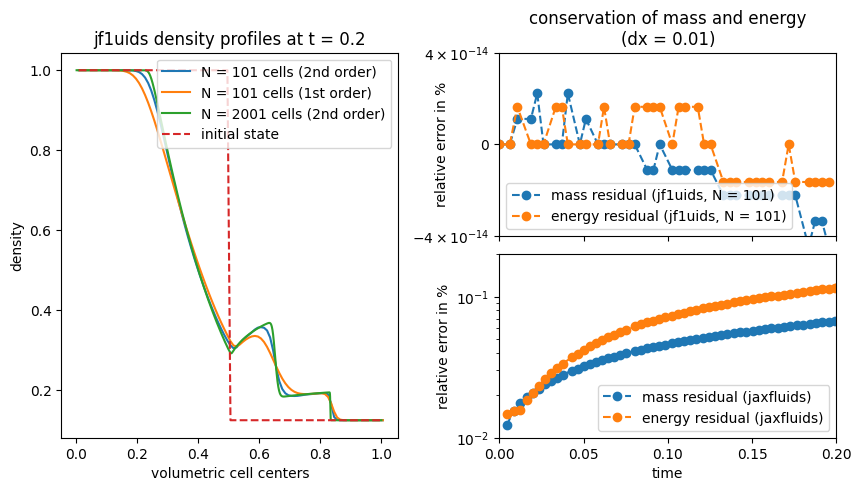
Visualization of the mass and energy development#
relative_mass_error = (checkpoints_shock100.total_mass - checkpoints_shock100.total_mass[0]) / checkpoints_shock100.total_mass[0] * 100
relative_energy_error = (checkpoints_shock100.total_energy - checkpoints_shock100.total_energy[0]) / checkpoints_shock100.total_energy[0] * 100
fig = plt.figure(figsize=(10, 5))
gs = GridSpec(2, 2, figure=fig)
gs.update(wspace=0.3, hspace=0.1)
ax1 = fig.add_subplot(gs[:, 0]) # Left plot spanning both rows
ax2 = fig.add_subplot(gs[0, 1]) # Top-right plot
ax3 = fig.add_subplot(gs[1, 1]) # Bottom-right plot
ax2.plot(checkpoints_shock100.time_points, relative_mass_error, "o--", label="mass residual (astronomix, N = 101)")
print(relative_mass_error)
ax3.set_xlabel("time")
ax2.legend(loc = "upper left")
ax2.set_ylabel("relative error in %")
ax2.yaxis.set_label_coords(-0.15, 0.5)
ax2.plot(checkpoints_shock100.time_points, relative_energy_error, "o--", label="energy residual (astronomix, N = 101)")
ax2.legend(loc = "lower left")
ax2.set_title("conservation of mass and energy\n(dx = 0.01)")
ax2.set_xticklabels([])
ax2.set_xlim(0, 0.2)
ax3.set_xlim(0, 0.2)
# less x ticks
ax3.set_xticks([0.0, 0.05, 0.1, 0.15, 0.2])
ax2.set_xticks([0.0, 0.05, 0.1, 0.15, 0.2])
sod_error = jnp.load("../data/sod_errors.npy")
times_jaxfluids, mass_errors_jaxfluids, energy_errors_jaxfluids = sod_error
ax3.plot(times_jaxfluids, mass_errors_jaxfluids, "o--", label="mass residual (jaxfluids)")
ax3.plot(times_jaxfluids, energy_errors_jaxfluids, "o--", label="energy residual (jaxfluids)")
ax3.set_ylim(1e-2, 0.2)
ax3.set_ylabel("relative error in %")
ax3.tick_params(axis='y')
ax3.set_yscale("log")
ax2.set_yscale("symlog")
ax2.set_ylim(-4e-14, 4e-14)
ax3.legend(loc = "lower right")
ax1.plot(helper_data_shock100.volumetric_centers, checkpoints_shock100.states[-1,0,:], label="N = 101 cells (2nd order)")
ax1.plot(helper_data_shock100_first_order.volumetric_centers, checkpoints_shock100_first_order.states[-1,0,:], label="N = 101 cells (1st order)")
ax1.plot(helper_data_shock.volumetric_centers, checkpoints_shock2000.states[-1,0,:], label="N = 2001 cells (2nd order)")
ax1.set_title("astronomix density profiles at t = 0.2")
# also plot the initial state in -- for reference
ax1.plot(helper_data_shock100.volumetric_centers, initial_state_shock100[0], "--", label="initial state")
ax1.set_xlabel("volumetric cell centers")
ax1.set_ylabel("density")
ax1.legend(loc = "upper right")
print(checkpoints_shock100.runtime)
print(checkpoints_shock2000.runtime)
plt.savefig("../figures/conservational_properties.png")
[ 0.00000000e+00 0.00000000e+00 1.11287917e-14 1.11287917e-14
2.22575833e-14 0.00000000e+00 0.00000000e+00 0.00000000e+00
2.22575833e-14 0.00000000e+00 1.11287917e-14 0.00000000e+00
0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
0.00000000e+00 -1.11287917e-14 -1.11287917e-14 0.00000000e+00
-1.11287917e-14 -1.11287917e-14 -1.11287917e-14 -1.11287917e-14
-1.11287917e-14 -1.11287917e-14 -2.22575833e-14 -2.22575833e-14
-2.22575833e-14 -2.22575833e-14 -2.22575833e-14 -2.22575833e-14
-2.22575833e-14 -2.22575833e-14 -2.22575833e-14 -2.22575833e-14
-4.45151666e-14 -3.33863750e-14 -3.33863750e-14 -4.45151666e-14]
0.0
0.0